Project members falling into the CTS4466 subclades can improve the data on this and the Pedigrees page through EXPERT LEVEL PARTICIPATION.
Subclade CTS4466 of R1b had at one time been called South Irish and was considered a distinctly Irish haplotype. That view has shifted somewhat. Project members willing to commit financially to doing more Y testing of SNPs should join the R-CTS4466 haplogroup project.
There are three known branches of HURLEY emerging under R-CTS4466 in the project. They are in private clusters 103, 108, and 120 respectively.
• CTS4466 > S1115 > A541 > S1121 > Z16252 > Z18170 > FGC29280
• CTS4466 > S1115 > A541 > S1121 > Z16252 > A9005 > FGC29068 > BY2880 > BY43744 > FGC17180 > A2224 > FTB79755
• CTS4466 > S1115 > A541 > A1135 > A195 > A761 > A88 > BY23574 > BY208692 > BY207561
A few members cite counties Cork and Kerry as geographic paternal line origins. Most do not give details.
The third SNP lineage shown above shows a descent from A88, thought to be a key mutation among descendants of the Ui Fidgenti.
CTS4466 members are encouraged to join both the R-CTS4466+ and Munster Irish projects if they have not already done so.
SNP Calendar / Timeline 
Click on those subclades marked with analysis for further detail on those Hurleys.
|
CTS4466 is positioned under FGC11134, and is, for the time being, its largest subclade discovered by far. CTS4466 has a dense presence in Southern Ireland, with numerous subclades appearing to emerge from that region. At the time of publication, some SNPs downstream from CTS4466 are not in ISOGG's or YFull's SNP tree or dated by YFull. Unfortunately, ISOGG is at least 2 years behind in updating long forms, which may change. There are also some difficulties aligning the data in FTDNA and in YFull. The three aforementioned subclades of interest under R-CTS4466 are lineages in the project and marked in the calendar. Compare these TMRCA dates to the emergence of surnames, starting around maybe 900 C.E. Many people acquired surnames from about then well into the first half of the second millennium. |
best viewed on wide screen computer
The buttons show or hide SNP information.
long forms
snps
tmrca
|
SNP Time and Geography Map 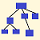
Back to Top
The locations pinned on the map are estimates, partly based on the genealogical data provided by project members, partly based on the location data at Scaled Innovation (Rob Spencer), and subject to some space limitations. Exact locations are not known. SNP formation date estimates are color coded based on Scaled Innovation.
Modal Haplotypes 
Back to Top
The first row of the table is an Irish Type II modal haplotype and is the REFERENCE ROW. It was borrowed from the Munster Irish Project. The remaining three rows are the modals of clusters 103, 108, and 120 in the Hurley project. They are marked to show their Genetic Distance from the reference row.
FGC29280 is explicitly shown both in the SNP calendar and on the Time and Geography map.
A2224 is shown on the SNP calendar. It is upstream from FTB79755, shown both on the SNP calendar and the Time and Geography map. The latter is a relatively recent mutation.
BY23574 is a parent SNP to BY208692; they are shown on the SNP calendar and the latter is on the Time and Geography map. BY208692 is upstream from BY207095 and BY207561, the latter two relatively recent mutations.
Each of these branches have multiple Hurley men in the project. The modals "roll them up" in to composite representations.
| Table #1 Modal Haplotyped of R-CTS4466 Hurleys | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| relative position | 1 | 2 | 3 | 4 | 5-6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14-15 | 16 | 17 | 18 | 19 | 20 | 21 | 22-25 | 26 | 27 | 28-29 | 30 | 31 | 32 | 33 | 34-35 | 36 | 37 | 38 | 39 | 40-41 | 42 | 43 | 44 | 45 | 46 | 47 | 48 | 49-50 | 51 | 52 | 53 | 54 | 55 | 56 | 57 | 58 | 59 | 60 | 61 | 62 | 63 | 64 | 65 | 66 | 67 | 68 | 69 | 70 | 71 | 72 | 73 | 74 | 75 | 76 | 77 | 78 | 79 | 80 | 81 | 82 | 83 | 84 | 85 | 86 | 87 | 88 | 89 | 90 | 91 | 92 | 93 | 94 | 95 | 96 | 97 | 98 | 99 | 100 | 101 | 102 | 103 | 104 | 105 | 106 | 107 | 108 | 109 | 110 | 111 | relative position | |||||||||
| Short Tandem Repeat Markers (STRS) | DYS393 | DYS390 | DYS19 | DYS391 | DYS385 | DYS426 | DYS388 | DYS439 | DYS389I | DYS392 | DYS389II | DYS458 | DYS459 | DYS455 | DYS454 | DYS447 | DYS437 | DYS448 | DYS449 | DYS464 | DYS460 | Y-GATA-H4 | YCAII | DYS456 | DYS607 | DYS576 | DYS570 | CDY | DYS442 | DYS438 | DYS531 | DYS578 | DYF395S1 | DYS590 | DYS537 | DYS641 | DYS472 | DYF406S1 | DYS511 | DYS425 | DYS413 | DYS557 | DYS594 | DYS436 | DYS490 | DYS534 | DYS450 | DYS444 | DYS481 | DYS520 | DYS446 | DYS617 | DYS568 | DYS487 | DYS572 | DYS640 | DYS492 | DYS565 | DYS710 | DYS485 | DYS632 | DYS495 | DYS540 | DYS714 | DYS716 | DYS717 | DYS505 | DYS556 | DYS549 | DYS589 | DYS522 | DYS494 | DYS533 | DYS636 | DYS575 | DYS638 | DYS462 | DYS452 | DYS445 | Y-GATA-A10 | DYS463 | DYS441 | Y-GGAAT-1B07 | DYS525 | DYS712 | DYS593 | DYS650 | DYS532 | DYS715 | DYS504 | DYS513 | DYS561 | DYS552 | DYS726 | DYS635 | DYS587 | DYS643 | DYS497 | DYS510 | DYS434 | DYS461 | DYS435 | Short Tandem Repeat Markers (STRS) | |||||||||
| member # |
|
member # | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| MODAL | 13 | 24 | 14 | 10 | 11-15 | 12 | 12 | 11 | 13 | 13 | 29 | 17 | 9-10 | 11 | 11 | 24 | 15 | 19 | 29 | 15-15-17-17 | 11 | 11 | 19-23 | 15 | 15 | 18 | 17 | 36-37 | 13 | 12 | 11 | 9 | 15-16 | 8 | 10 | 10 | 8 | 10 | 10 | 12 | 23-23 | 16 | 10 | 12 | 12 | 15 | 8 | 12 | 22 | 20 | 13 | 12 | 11 | 13 | 11 | 11 | 12 | 11 | 35 | 15 | 9 | 16 | 12 | 25 | 26 | 19 | 12 | 11 | 13 | 12 | 11 | 9 | 13 | 11 | 10 | 11 | 11 | 30 | 12 | 13 | 24 | 13 | 10 | 10 | 21 | 15 | 19 | 14 | 24 | 16 | 12 | 15 | 24 | 12 | 24 | 18 | 10 | 14 | 18 | 9 | 12 | 11 | MODAL | |||||||||
| FGC29280-MOD | 13 | 24 | 14 | 10 | 11-15 | 12 | 12 | 11 | 13 | 13 | 29 | 17 | 9-10 | 11 | 11 | 24 | 15 | 19 | 29 | 15-15-17-17 | 11 | 11 | 19-23 | 15 | 15 | 18 | 17 | 36-37 | 13 | 12 | 11 | 9 | 15-16 | 8 | 10 | 10 | 8 | 10 | 10 | 12 | 23-23 | 17 | 10 | 12 | 12 | 15 | 8 | 12 | 22 | 20 | 13 | 12 | 11 | 13 | 11 | 11 | 12 | 11 | 35 | 15 | 9 | 16 | 12 | 25 | 26 | 19 | 12 | 11 | 13 | 12 | 11 | 9 | 13 | 11 | 10 | 11 | 11 | 30 | 12 | 13 | 24 | 13 | 10 | 11 | 21 | 15 | 18 | 14 | 24 | 16 | 12 | 15 | 24 | 12 | 24 | 18 | 10 | 14 | 18 | 9 | 12 | 11 | FGC29280-MOD | |||||||||
| A2224-MOD | 13 | 24 | 14 | 10 | 11-15 | 12 | 12 | 11 | 13 | 13 | 29 | 17 | 9-10 | 11 | 11 | 24 | 15 | 19 | 29 | 15-15-17-18 | 11 | 11 | 19-23 | 16 | 15 | 18 | 17 | 36-38 | 13 | 12 | 11 | 9 | 15-16 | 8 | 10 | 10 | 8 | 10 | 10 | 12 | 23-23 | 17 | 10 | 12 | 12 | 15 | 8 | 13 | 22 | 20 | 13 | 12 | 11 | 14 | 11 | 11 | 12 | 11 | 35 | 15 | 9 | 16 | 12 | 25 | 26 | 19 | 12 | 11 | 13 | 12 | 11 | 9 | 13 | 11 | 10 | 11 | 11 | 30 | 12 | 13 | 24 | 13 | 10 | 10 | 20 | 15 | 18 | 14 | 24 | 16 | 12 | 15 | 25 | 12 | 24 | 18 | 10 | 14 | 18 | 9 | 12 | 11 | A2224-MOD | |||||||||
| BY23574-MOD | 13 | 24 | 14 | 10 | 11-15 | 12 | 12 | 11 | 13 | 14 | 29 | 18 | 9-10 | 11 | 11 | 24 | 15 | 19 | 29 | 15-15-17-18 | 11 | 11 | 19-23 | 15 | 15 | 18 | 16 | 36-37 | 13 | 12 | 11 | 9 | 15-16 | 8 | 10 | 10 | 8 | 10 | 10 | 12 | 23-23 | 16 | 10 | 12 | 12 | 15 | 8 | 12 | 22 | 20 | 13 | 12 | 11 | 13 | 11 | 11 | 12 | 11 | 35 | 15 | 9 | 16 | 12 | 25 | 26 | 19 | 12 | 11 | 13 | 12 | 11 | 9 | 12 | 11 | 10 | 11 | 11 | 30 | 12 | 13 | 24 | 13 | 10 | 10 | 21 | 15 | 19 | 14 | 25 | 16 | 12 | 15 | 24 | 12 | 24 | 18 | 10 | 14 | 18 | 9 | 12 | 11 | BY23574-MOD | |||||||||
| member # |
| member # | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
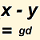 Genetic Distances of Hurley R-CTS4466 Modals to Irish Type II Modal
Genetic Distances of Hurley R-CTS4466 Modals to Irish Type II Modal
Back to Top
See Genetic Distance in Page Help for details. Note the remarks on the reliability of TMRCA estimation based on STR markers gradually deteriorating as we go further back in time. This would be particularly applicable to modal comparison, as opposed to comparison of individuals.
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
TMRCA Calculations of Hurley R-CTS4466 Modals 

Back to Top
See Time to Most Common Recent Ancestor in Page Help for details. Note the remarks on the reliability of TMRCA estimation based on STR markers gradually deteriorating as we go further back in time. This would be particularly applicable to modal comparison, as opposed to comparison of individuals.
|
|||||||||||||||||||||||||||||||||||||||||
| Hurley, Harley, Herilihy at FTDNA |
DNA Portal | Records | Project Pedigrees |
| See ABOUT for website information. | |||
| Copyright © 2018-2022 hurley.dnagen.org. All rights reserved. | |||